
- 4PEAKS DOWNLAD INSTALL
- 4PEAKS DOWNLAD SOFTWARE
- 4PEAKS DOWNLAD CODE
- 4PEAKS DOWNLAD FREE
- 4PEAKS DOWNLAD WINDOWS
SeqTrace has modest hardware requirements: computers capable of running modern graphical desktop operating systems should also be adequate for processing trace files with SeqTrace.
4PEAKS DOWNLAD WINDOWS
SeqTrace is distributed as a binary package for Windows and as a source package for all other operating systems. The SeqTrace program, source code, and supporting documentation are all freely available at.
4PEAKS DOWNLAD SOFTWARE
Additionally, PyInstaller ( ) was used to create a self-contained SeqTrace package for Microsoft Windows that eliminates all software dependencies on that platform.
SeqTrace was tested on several popular GNU/Linux distributions (Xubuntu 11.10, Ubuntu 11.04, and Linux Mint 12), recent versions of Microsoft Windows (XP, Vista, and 7), and Apple OS X 10.6 in the native and X11 environments. Consequently, trace files processed with SeqTrace are expected to include quality scores, and SeqTrace does not attempt to compute them if they are absent. 7, 8 However, all modern capillary sequencing instruments use base-calling software that calculate quality scores, so virtually all recent chromatogram trace files include them. Calculating accurate quality scores is a complex problem, due in part to variations in sequencing machines and techniques.
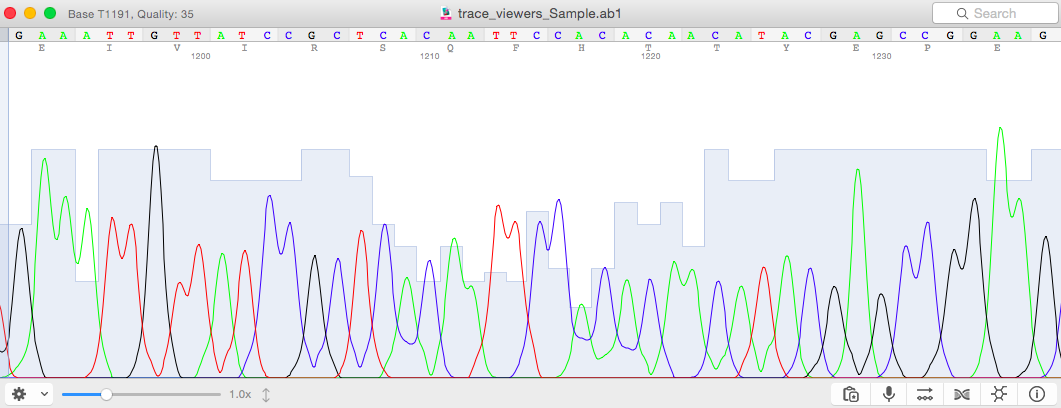
For matched forward and reverse sequences, base mismatches and gaps both represent sequencing errors and should be weighted equally.Īccurate base-call quality scores are essential for producing finished sequences.
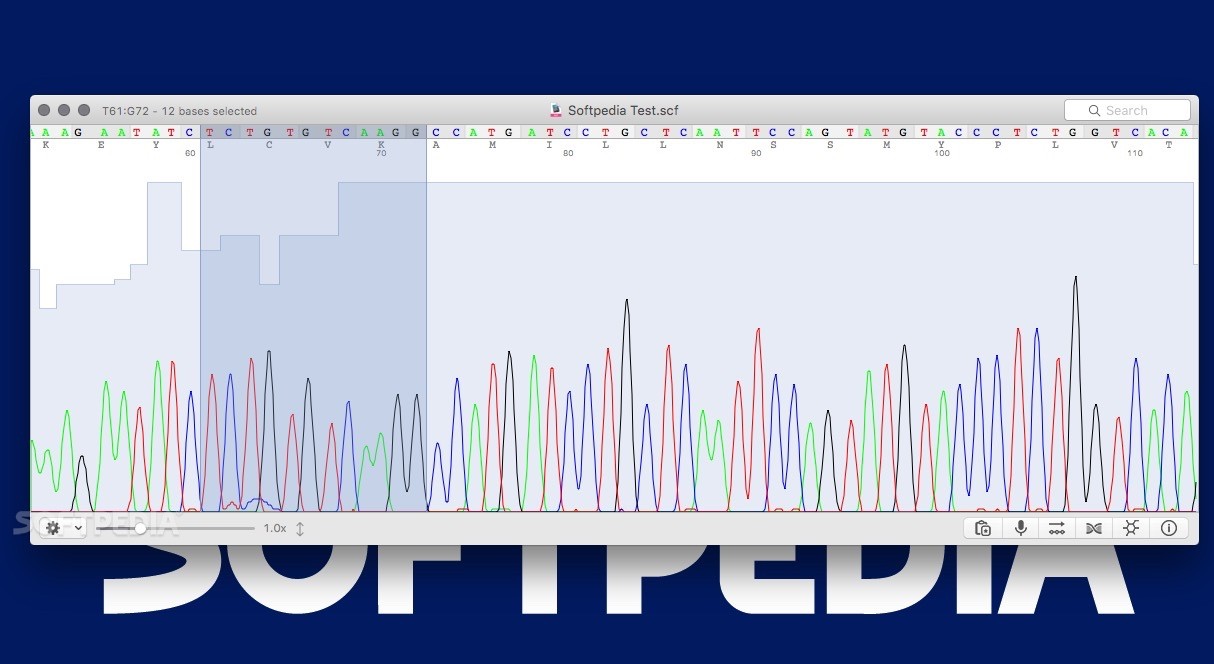
5, 6 Base mismatches, interior gap openings, and gap extensions are all given the same penalty (i.e., linear gap penalties are used). To accomplish this, SeqTrace uses a customized Needleman-Wunsch pairwise alignment algorithm. Generating a consensus sequence from matching forward and reverse sequencing reads requires first computing a pairwise global alignment of the raw forward and reverse sequences.
4PEAKS DOWNLAD INSTALL
To support multiple input and ouput file formats without requiring the user to install additional software libraries, all file formats were implemented directly in Python as part of the SeqTrace application. To meet these requirements, SeqTrace follows object-oriented design principles and was implemented in Python ( ) using the cross-platform GTK+ windowing toolkit ( ).
4PEAKS DOWNLAD CODE
A secondary goal was to ensure that the SeqTrace source code could be reused easily in other bioinformatics applications. SeqTrace was designed to be a graphical, user-friendly software program that would run on most of the common operating systems in current use.
4PEAKS DOWNLAD FREE
SeqTrace is free and open-source software that runs on all popular operating systems. Although SeqTrace was designed with batch processing in mind, it can also serve as a general-purpose trace viewer and editor. SeqTrace is intended specifically for sequencing projects that require converting trace files directly into high-quality, finished sequences, and it provides a graphical, user-friendly interface for automating the entire process. SeqTrace, a new computer program described in this communication, was created to help fill this gap. Although commercial software is available to handle these tasks, free software options are generally much more limited. If done manually, these steps can be very time consuming, especially for large projects. Paired forward and reverse reads of PCR products are also frequently used to ensure final sequence quality this requires aligning the forward and reverse sequences and determining a single consensus sequence from the pair. At a minimum, this involves inspecting each trace file to identify problematic sequencing runs, remove unreliable base calls, and trim the ends of the sequence. Modern Sanger sequencing instruments, however, generate “raw” chromatogram trace files that require further processing to obtain sequences of sufficient quality for downstream analyses. Many such projects require high-quality sequencing reads from a relatively large number of PCR amplicons. 2 Although high-coverage, high-volume sequencing has largely moved to “next-generation” technologies, Sanger sequencing remains a popular and indispensable tool for low-coverage sequencing applications, such as phylogenetic analyses or DNA barcoding efforts. Since its development in the late 1970s, Sanger chain-termination DNA sequencing 1 has become a widely used, essential technique of molecular biology.


 0 kommentar(er)
0 kommentar(er)
